chip seq venn diagram
Batch annotation of the peaks identified from either ChIP-seq ChIP-chip. Transcription factors have two major.
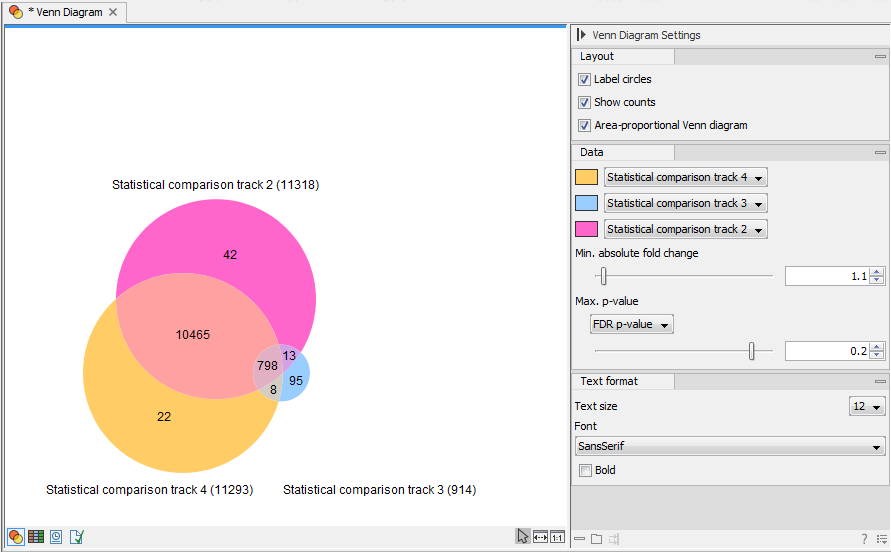
Clc Manuals Clcsupport Com
Examination of ChIP-seq and seq-DAP-seq bound regions and DE genes in the fourth whorl during carpel development reveals a high degree of overlap Figure 5 with not only many well.

. Transcription factors have two. Transcription factors have two. The Venn diagram on the right shows results from RNA-seq and ChiP-seq experiments designed to find Pax6 target genes as we discussed in class.
A list of target genes generated from ChIP. We performed ChIP-seq analyses to explore genomic regions. The Venn diagram on the right shows results from RNA-seq and ChiP-seq experiments designed to find Pax6 target genes as we discussed in class.
Workflow for ChIP-seq experiments of single transcription factor with replicates. The first part of ChIP-seq analysis workflow consists in read preprocessing. Workflow for ChIP-seq experiments of single transcription factor with replicates.
A Venn diagram of ChIP-seq peaks. LibraryVennDiagram Loading required package. Call peaks with at least 2-3 software tools such as MACS2 slice coverage calling Bioc PeakSeq F-Seq Homer ChIPseqR or CSAR.
Once peaks and TF binding sites are identified in ChIP-Seq data peaks can be annotated to the genome to find the nearest transcription start site. 51 Calculate overlap with genomic features like introns exons. This workflow shows how to convert BEDGFF files to GRanges find overlapping peaks.
The Venn diagram is not dawn to scale. We draw this as a Venn diagram using the drawpairwisevenn function from the VennDiagram package. Contribute to ragakChIP-seq development by creating an account on GitHub.
B Read densities over ChIP-seq peaks of C11orf95fus1 and C11orf95-RELAfus1. 5 Functional annotation of ChIP-seq peaks. Make Venn Diagram from two or more peak ranges Also calculate p-value to determine.
44 Make a Venn-diagram showing the overlap of of ER and FOXA1 peaks. 11 rows In ChIPpeakAnno. Contribute to Lie-neChIP-seq development by creating an account on GitHub.
Compare the results with peaks identified.
Utah State University Bioinformatics Facility
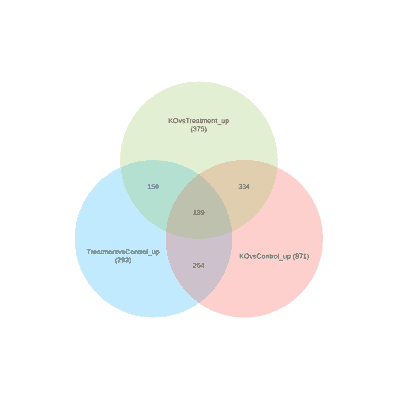
Gene Venn Diagram Tool Generate With Omicsbox

Bend3 Safeguards Pluripotency By Repressing Differentiation Associated Genes Pnas
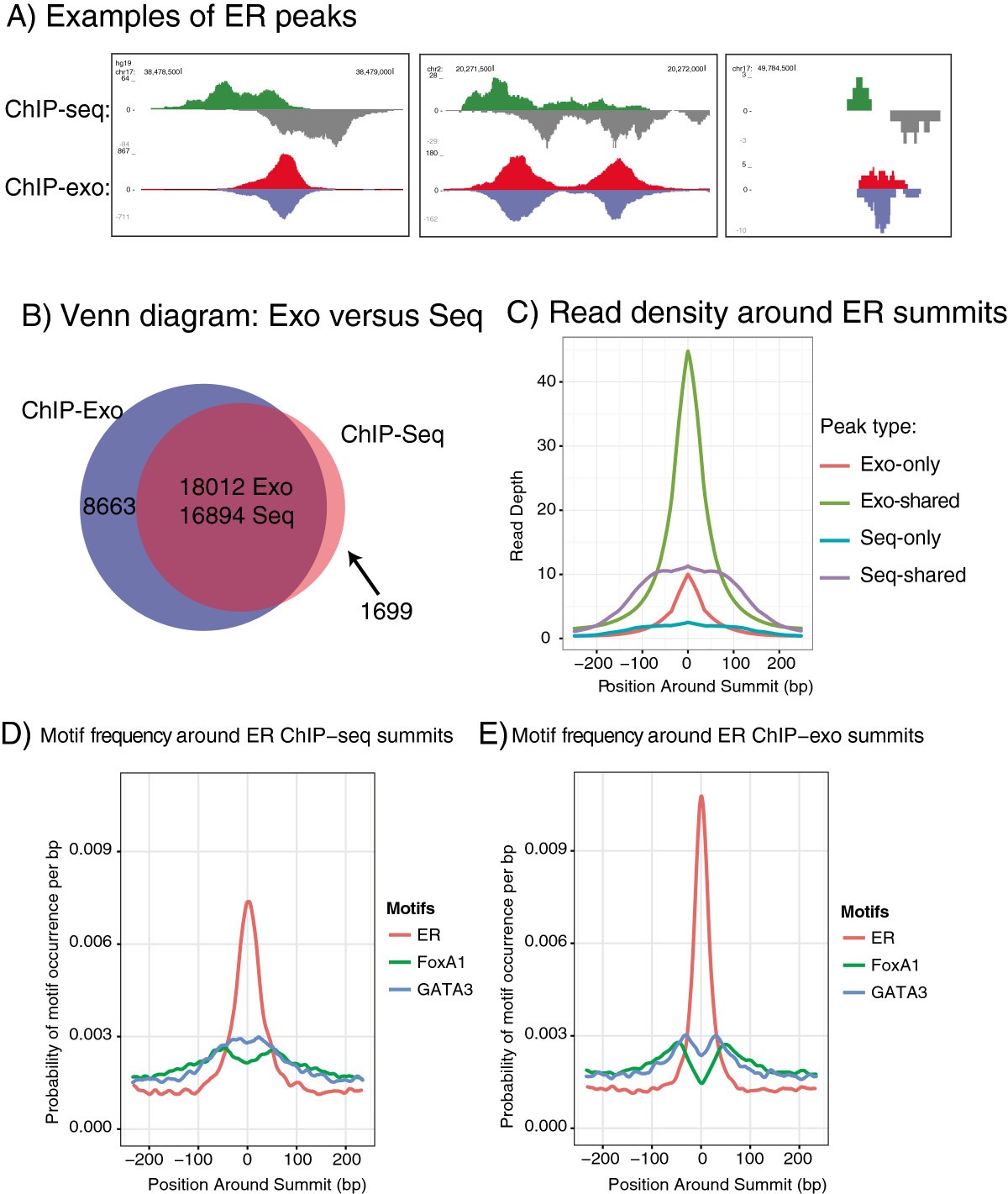
Development Of An Illumina Based Chip Exonuclease Method Provides Insight Into Foxa1 Dna Binding Properties Genome Biology Full Text
Loss Of The Caenorhabditis Elegans Pocket Protein Lin 35 Reveals Muvb S Innate Function As The Repressor Of Dream Target Genes Plos Genetics

7 Rna Seq Data Ansysis Basic Scatter Plot Venn Diagram Etc Youtube
Histone Modifier Gene Mutations In Peripheral T Cell Lymphoma Not Otherwise Specified Haematologica
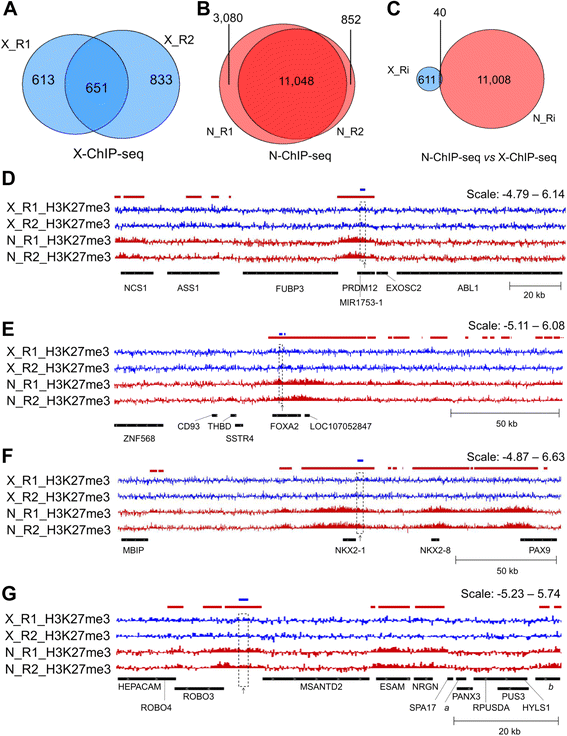
An Assessment Of Fixed And Native Chromatin Preparation Methods To Study Histone Post Translational Modifications At A Whole Genome Scale In Skeletal Muscle Tissue Biological Procedures Online Full Text
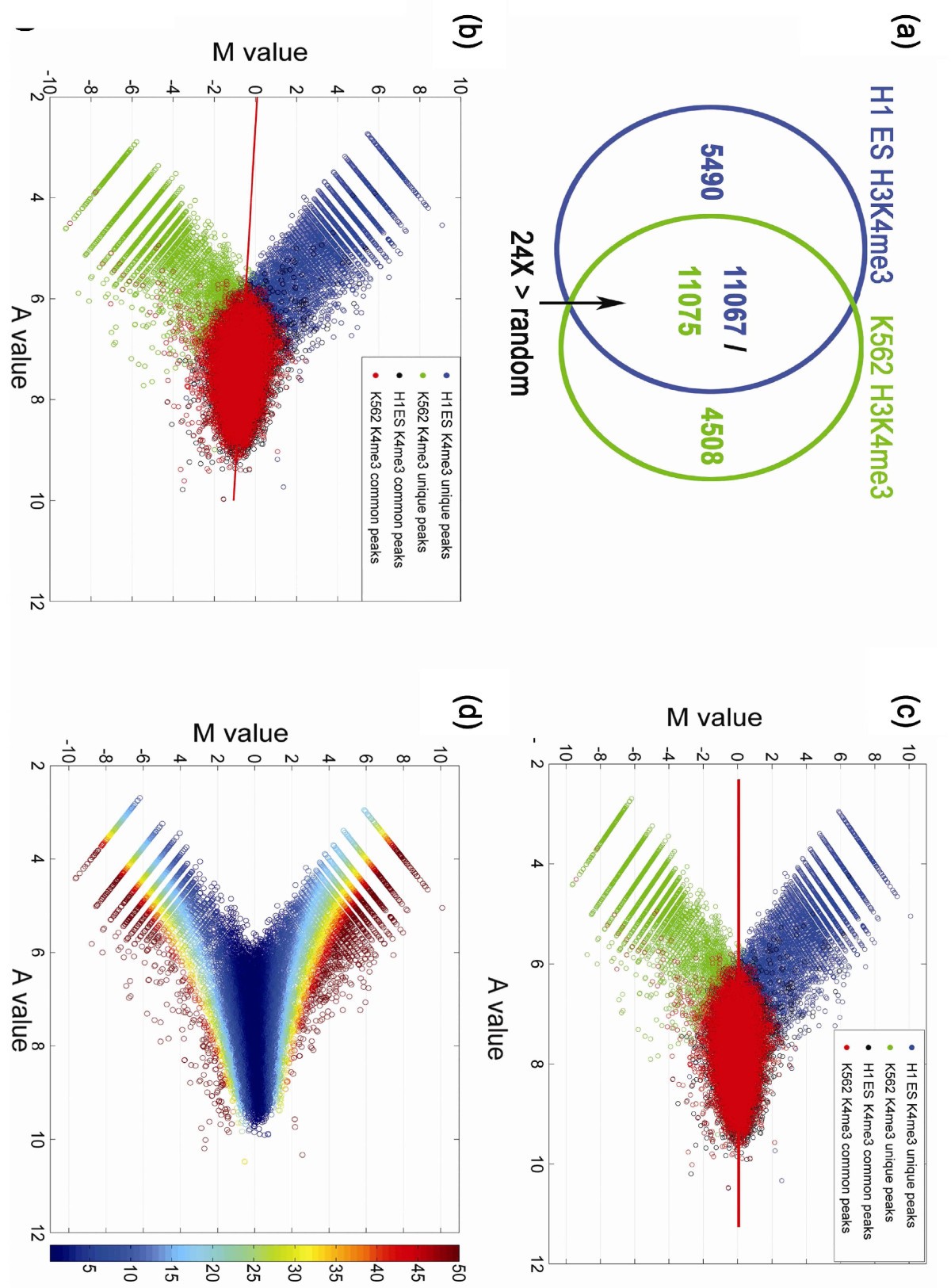
Manorm A Robust Model For Quantitative Comparison Of Chip Seq Data Sets Genome Biology Full Text
Srplot Free Online Genomic Peaks Overlap Venn Diagram
Cistrome Cancer
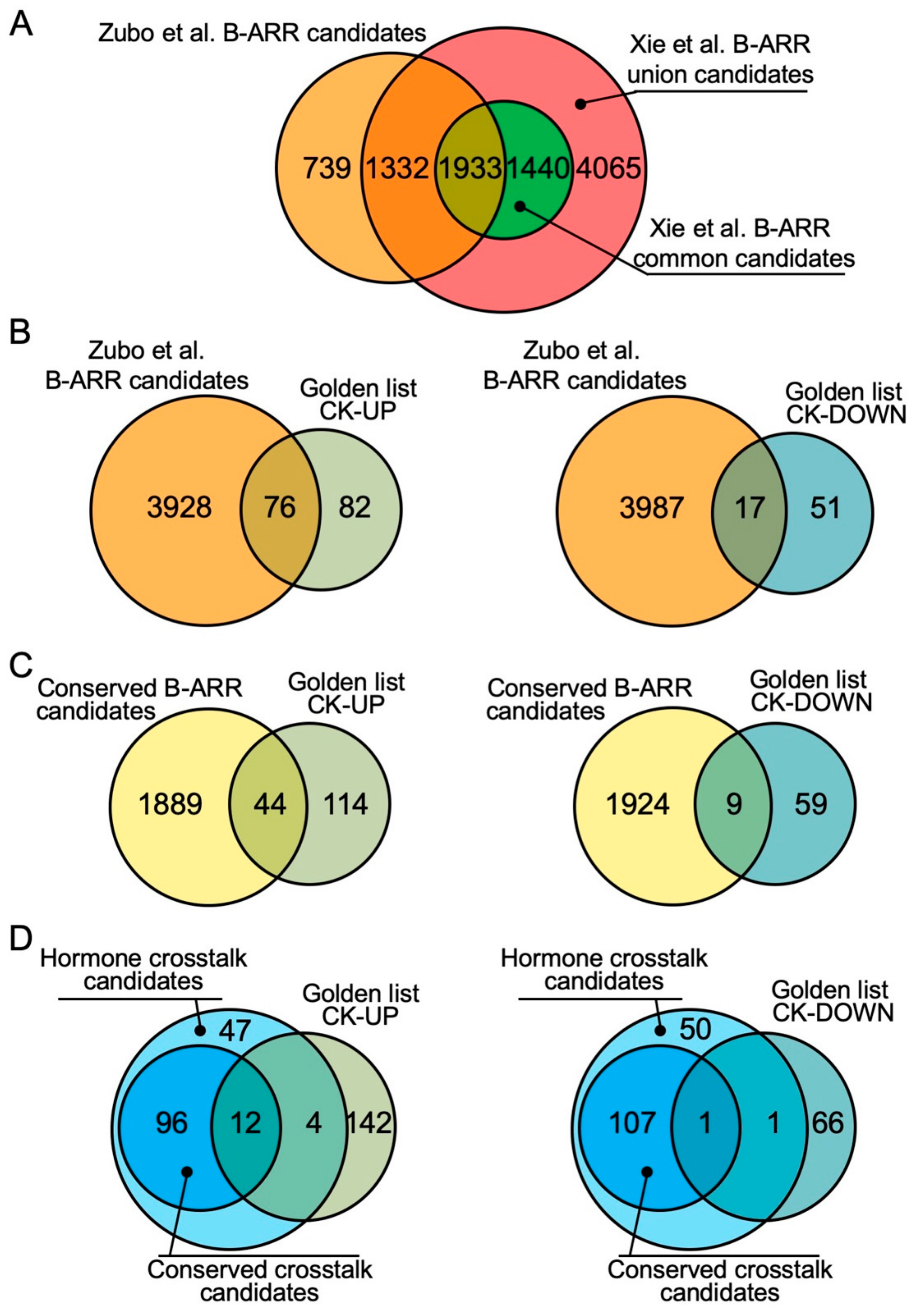
Plants Free Full Text Role Of The Cytokinin Activated Type B Response Regulators In Hormone Crosstalk Html
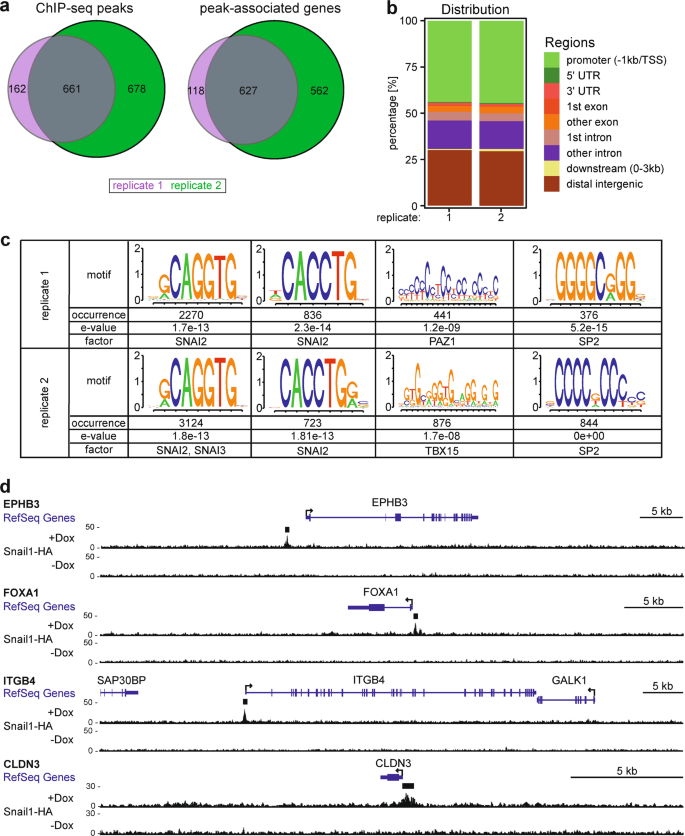
Genome Wide Mapping Of Dna Binding Sites Identifies Stemness Related Genes As Directly Repressed Targets Of Snail1 In Colorectal Cancer Cells Oncogene
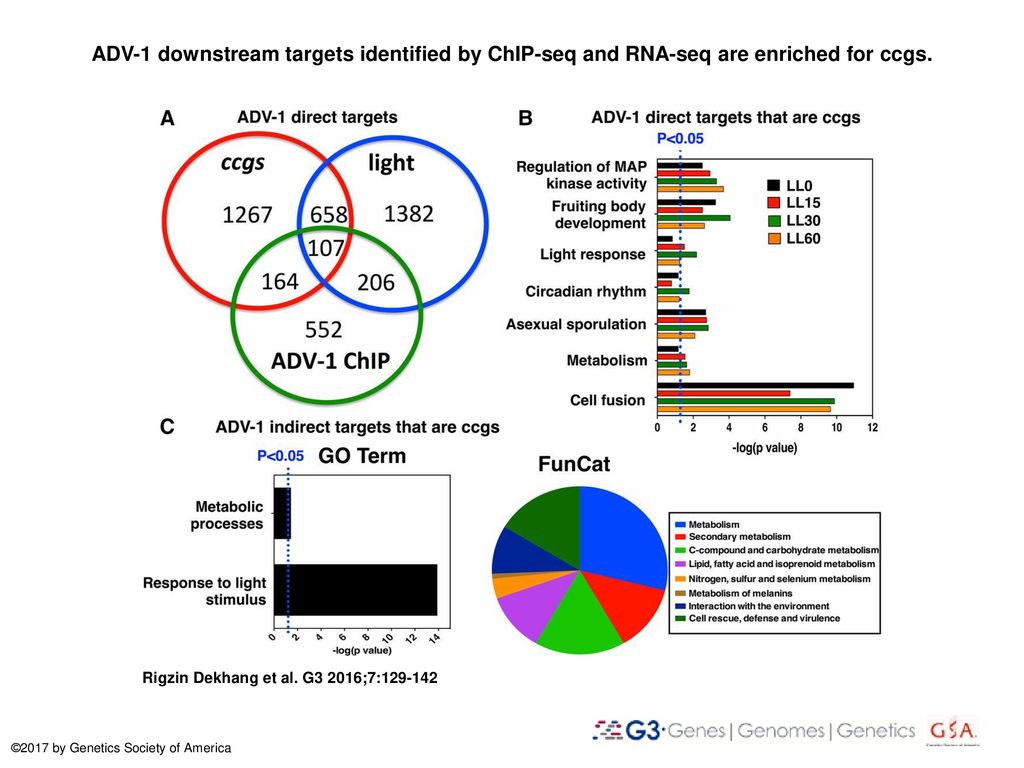
Adv 1 Downstream Targets Identified By Chip Seq And Rna Seq Are Enriched For Ccgs Adv 1 Downstream Targets Identified By Chip Seq And Rna Seq Are Enriched Ppt Download
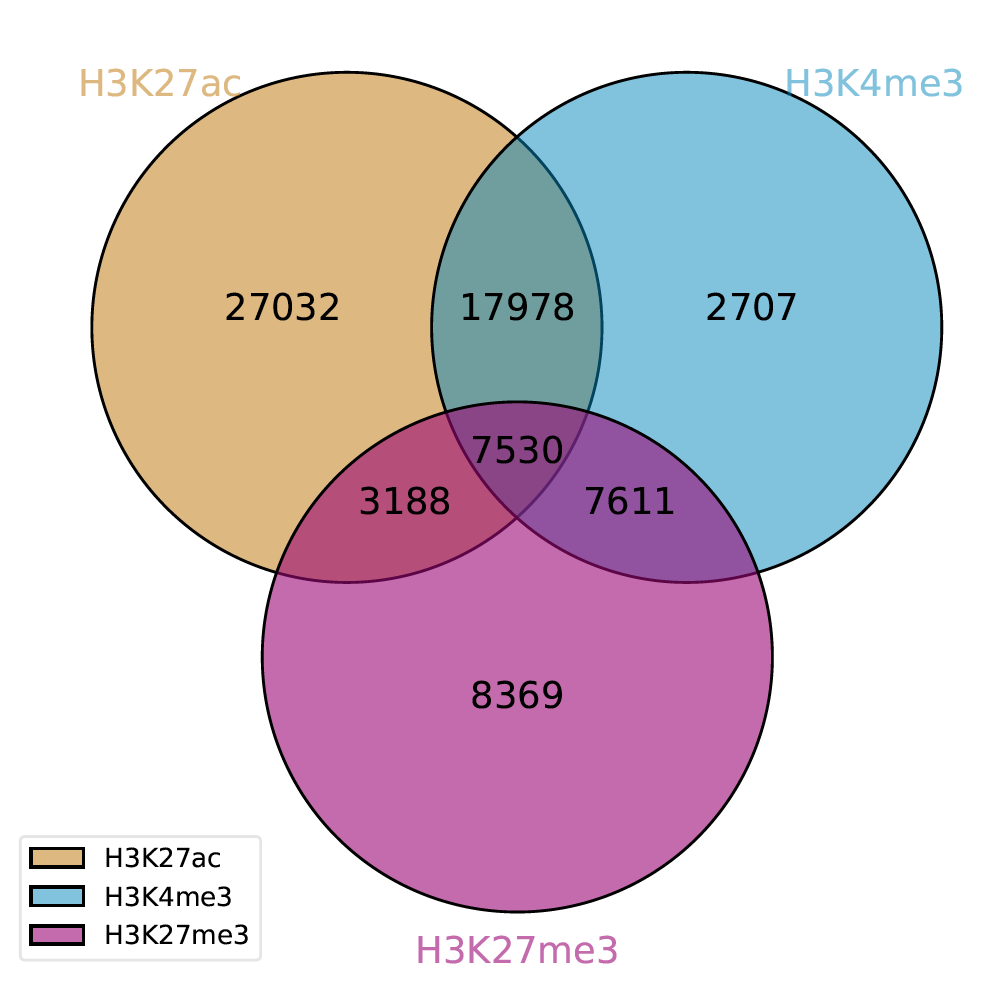
Example Gallery A Tool For Intersection And Visualization Of Multiple Gene Or Genomic Region Sets

Intervene A Tool For Intersection And Visualization Of Multiple Gene Or Genomic Region Sets Biorxiv

Venn Diagram Of Intersections Of Rna Seq And Microarray Data Of Post Noise Filtered Genes Representing 7 506 Microarray And 9 228 Rna Seq Genes